The Gerold Lab | Cellular Virology
Our research group aims to understand the underlying biochemical principles of an infection of host cells by viruses. The main focus of our studies is the investigation of viral entry and replication in host cells. We investigate protein interactions during virus life cycle by using high-resolution quantitative protein analysis. In recent years, we have characterized protein interactions during infection with chronic hepatitis C virus (HCV) and emerging insect-borne viruses. These studies have also been extended to SARS-CoV-2. In the long term, the knowledge about host factors can be used for to develop anti-infectives and can help understand tissue and host tropism of viruses.
Emergence of viruses represents a global challenge that is accelerated by anthropogenic changes to ecosystems. Virus-transmitting mosquito species were able to spread to Europe as a result of climate change, for instance. Using mosquito-borne viruses, such as the chikungunya virus, and emerging viruses, such as SARS-CoV-2, we focus on protein networks involved in viral infection. For this purpose, we are using proteomics techniques. Our work aims to identify similarities and differences in host factor usage of zoonotic viruses, i.e. viruses that are transmitted between animals and humans. This will open new avenues for the development of antiviral drugs and the understanding of host and tissue tropism, which will ultimately contribute to risk assessment of transmission routes, disease progression and the pandemic potential of viruses.
Our group is interested in understanding the protein and lipid determinants of viral infections. The figure to the right shows a model of the chikungunya virus replication complex at the plasma membrane with the viral genome in the centre and host proteins and lipids we have identified embedded in the membrane. The dodecameric ring at the neck represents the non-structural protein 1 (nsP1) of the chikungunya virus. Details can be found in Lasswitz et al., mBio 2022.
Projects
Understanding Fundamental Mechanisms Governing Insect Cell Membrane Deformability
Duration:
October 2023 – September 2026
Details:
Insect cell membranes differ from mammalian membranes in deformability, lipid content and distribution. Enveloped viruses transmitted by insects require close interactions with cell membranes to enter cells, replicate their genomes in cells and exit cells. Despite fundamental biophysical differences between insect and mammalian membranes, viruses can productively infect cells from both phyla. Decades of studies on insect viruses have not investigated the mechanism of insect membrane deformation and its exploitation by viruses. Our newly formed team works at the interface between insect genetics, biophysics and infection biology and has assembled innovative technologies to break through this barrier. Through the PIs’ combined expertise, we will: 1. use gene editing libraries in insect cells to identify host factors that control virus-membrane interactions; 2. visualize membrane deformation at high resolution; 3. assess the impact of membrane deformation on permeability to insect-borne viruses. These studies will provide insight into the mechanisms that control insect membrane shape and revolutionize our understanding of virus adaptation in fundamentally different species.
Funding:
The International Human Frontier Science Program Organization (HFSP), 1.106.000 EUR
The Role of Tetraspanins in Cross-Species Transmission of Re-emerging Alphaviruses
Duration:
April 2022 – March 2025
Details:
Tetraspanins are evolutionarily conserved integral membrane proteins with a length of 200-350 amino acids. Through their large extracellular loop, they mediate protein-protein and protein-lipid interactions in cell membranes, forming membrane microdomains known as “tetraspanin networks”. In humans and mice, 33 tetraspanins have been described, and mosquito species express at least 15 tetraspanin orthologs. In mammalian cells, tetraspanins are host cofactors for various viruses, including papillomaviruses, influenza viruses, hepatitis C viruses, HIV-1 and coronaviruses (Gerold et al, 2015; Bruening et al, 2018; Banse et al, 2018; Alberione et al, 2020; Palor et al, 2020). For hepatitis C virus, colleagues and we have shown that the tetraspanin CD81 is a factor that determines host range (Vogt et al. 2013; Scull et al., 2015; von Schaewen et al., 2016).
The proposed project aims to characterize in detail which of the 33 human tetraspanins besides CD81 are host factors for alphaviruses and whether tetraspanins from reservoir species, dead-end host species and transmitting mosquito vectors serve as host factors of alphaviruses. Thus, the work will contribute to the understanding of the molecular composition and function of alphavirus replication complexes and determine the role of tetraspanins in the species spectrum, transmission and consequently the emergence of alphaviruses.
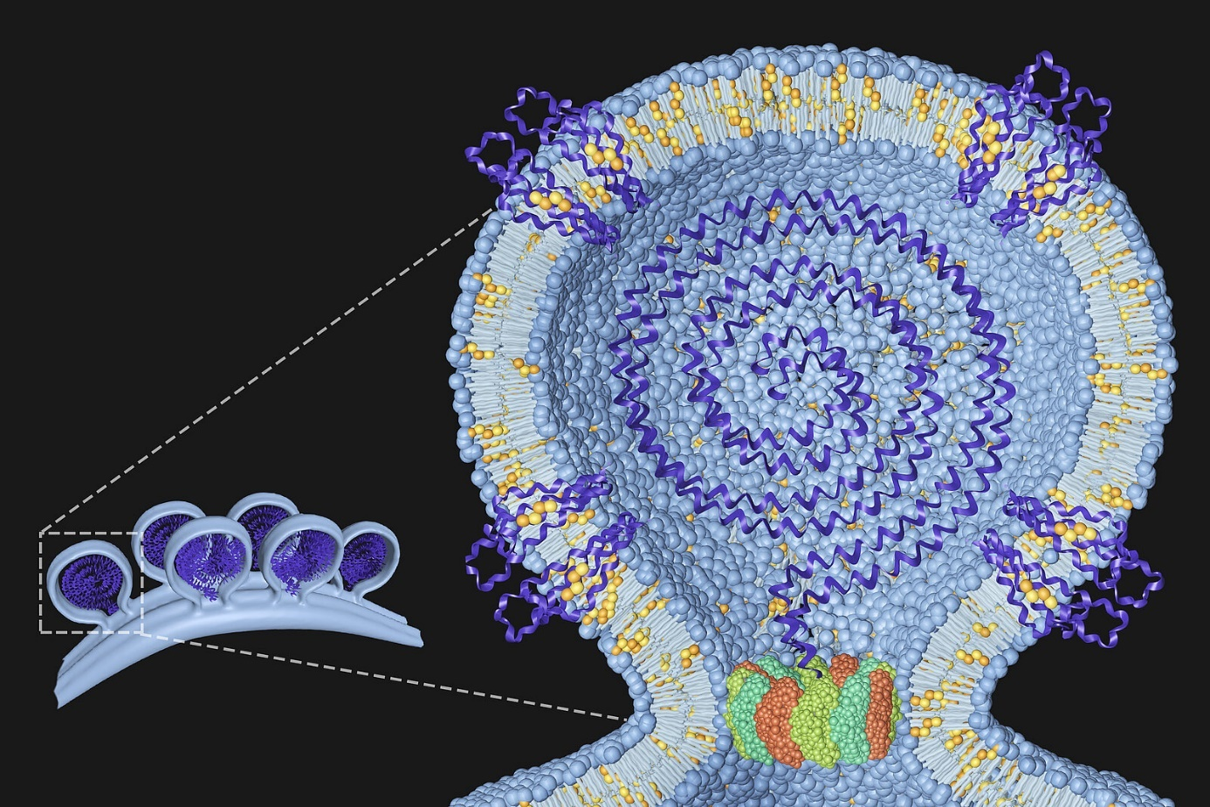
Our group is interested in understanding the protein and lipid determinants of viral infections. The figure shows a model of the chikungunya virus replication complex at the plasma membrane with the viral genome in the centre and host proteins and lipids we have identified embedded in the membrane. The dodecameric ring at the neck represents the non-structural protein 1 (nsP1) of the chikungunya virus. Details can be found in Lasswitz et al., mBio 2022.
Funding:
Deutsche Forschungsgesellschaft (DFG), 110.700 EUR
Development of an Ex Vivo Model to Study the Zoonotic Transmission of Noroviruses
Duration:
May 2023 – October 2026
Details:
Noroviruses are the most common viral cause of acute gastroenteritis. At least 40 noroviruses have been described and new variants appear regularly. The development, emergence and spread of noroviruses are not fully understood, in particular the role of a potential animal reservoir has not been fully investigated. There are several indications of norovirus transmission between humans and animals. However, so far there is no in vitro system to study transmission between different species. In this project, we aim to develop an ex-vivo system based on intestinal biopsies from different animal species to experimentally study norovirus transmission between different species. We will use biopsies from dogs, pigs and chickens, as they occur in large numbers and are in close contact with humans. The risk of transmission is therefore increased. Precision-cut intestinal sections will be established and used to study norovirus binding, invasion/internalisation and replication in the respective host tissues. In addition, the role of known susceptibility factors, the histo-blood group antigens, will be investigated. Once this explant system is established, it will allow the study of norovirus receptors, attachment factors and other host factors in humans and non-human animal species. The system will also be useful for the study of other enteric viruses with zoonotic potential, including coronaviruses, influenza viruses and canine and porcine noroviruses.
Funding:
BMBF, 198.361 EUR
Model for Arbovirus Infection of the Skin - MOZART
Duration:
October 2023 – July 2025
Details:
Every year, around 750,000 people die from mosquito-borne diseases, including malaria, dengue fever, Rift Valley fever and Chikungunya fever. The incidence of these diseases is expected to increase significantly in the coming decades as the distribution areas of several mosquito species expand due to climate change.
Mosquitoes not only transmit diseases, but can also influence the severity of the diseases they transmit. Experimental infections of animals have shown that the transmission of arboviruses through mosquito bites can lead to an increase in the severity of the disease compared to an artificial infection. It is also known that the saliva of insects and ticks can promote the progression of vector-borne diseases. The skin is the first organ to be exposed to an arbovirus infection, so the initial infection events also significantly determine the course of the disease. It is therefore essential for many research questions to investigate a natural infection of the skin via a mosquito bite.
Until now, this could only be achieved through in-vivo experiments (animal experiments). The aim of this project is to develop an alternative by replacing animal models with human skin explants for ethical and physiological reasons. The challenge of this study will be to establish an assay in which mosquitoes recognise the skin explants as potential hosts. The aim is for mosquitoes to successfully bite the skin explants. As an example, in a second step Rift Valley Fever Virus (RVFV) and/or Chikungunya Virus (CHIKV)-infected mosquitoes will infect the skin explants via a bite. If successful, the project results will make it possible to select from various models for future research questions.
Funding:
BMBF, 198.361 EUR
Identification and Characterization of Alphavirus Host Factors Determining Human Tissue Tropism
Duration:
January 2022 – December 2025
Details:
New and re-emerging viruses pose a serious health problem. In particular, mosquito-borne infectious diseases are on the rise as certain mosquito species invade new geographic regions. Alpha viruses such as Chikungunya virus (CHIKV) and Venezuelan equine encephalitis virus (VEEV) belong to the group of mosquito-borne viruses. CHIKV causes long-lasting arthritis symptoms, while VEEV causes meningitis. Why the two closely related viruses cause disease symptoms in different tissues, i.e. joints versus the central nervous system, is largely unknown. This knowledge gap is reflected in an insufficient understanding of host factors of CHIKV and VEEV. In this research project, we build on our findings that the phosphatidylserine (PS) receptor TIM-1 (T cell immunoglobulin mucin receptor) and a tetraspanin are host factors of CHIKV. We hypothesise that (a) VEEV also uses PS receptors and tetraspanins as host factors, (b) TIM-1 and the tetraspanin interact with other proteins that promote infection and (c) that some of these proteins contribute to the tissue tropism of CHIKV and VEEV. To test our hypothesis, we will first apply the latest quantitative proteomics techniques to identify entry factors and receptors of CHIKV and VEEV. We will then test the role of PS receptors and the 33 human tetraspanins in VEEV infection using RNA interference. TIM-1 and tetraspanin associated proteins will be determined by proximity labelling, high-resolution affinity enrichment and mass spectrometry. Finally, we will analyse the contribution of the identified host factors to the tissue tropism of CHIKV and VEEV by proteomic analysis, single cell sequencing and virological methods. In particular, skin organoids will be infected and the viral target cells in the skin as well as host factor expression will be determined. In addition, we will integrate expression data from databases and total cell proteome measurements of fibroblasts and neuronal cells into the data set. Using the latest bioinformatic analyses (feature selection), we will then determine host factors that are highly likely to contribute to tissue tropism and the different disease patterns. These host factors will be systematically knocked out in relevant human cells and finally in mice to elucidate their contribution to the disease pattern in vitro and in vivo. In summary, this research programme aims to provide mechanistic insights into the infection process of two important human pathogens. The results will lead to a better understanding of the symptoms caused by CHIKV and VEEV and may provide starting points for therapies to alleviate or cure chikungunya fever and Venezuelan encephalitis.
Funding:
Deutsche Forschungsgesellschaft (DFG), 398.333 EUR
Understanding the Role of Phosphatidylserines and its Receptors in Cross-Species Transmission of Alphaviruses
Duration:
October 2021 – June 2025
Details:
Alphaviruses (family Togaviridae) are emerging and re-emerging small enveloped RNA viruses that are transmitted from animal reservoirs to humans by mosquitoes and can cause paralysing joint pain or encephalitis. Depending on the reservoir species and the transmitting mosquito vector, they can be found in different geographical regions. While the Chikungunya virus (CHIKV) and the O’nyong’nyong virus (ONNV) were historically restricted to tropical and subtropical climate zones, the Sindbis virus (SINV) and the Ross River virus (RRV) are mainly found in Scandinavia and Australia respectively. The adaptation of CHIKV to new mosquito vectors and global warming have led to the emergence of the virus in Europe and made it a potential public health problem in Germany and neighbouring countries. An introduction of the Venezuelan
equine encephalitis virus (VEEV), which circulates in the Americas and causes neurological symptoms in equids and humans, is also possible in the future. To date, there are no human vaccines or antiviral drugs against arthritogenic and neurotropic alphaviruses on the market. This is due to the fact that the molecular mechanisms of the infection process and the critical host factors involved in alphavirus infection and cross-species transmission are not known. Recently, our team found that the phosphatidylserine receptor T-cell immunoglobulin and mucin domain 1 (TIM-1) is a CHIKV attachment factor. Another prominent group of phosphatidylserine receptors is the Tyro3, AXL and MerTK (TAM) receptor tyrosine kinase (RTK) family, which comprises the three proteins Tyro3, AXL and MerTK. The physiological function of the TIM and TAM receptors is to bind and internalise apoptotic bodies that release phosphatidylserine on the outer membrane leaflet. We were able to show that TIM-1, but not AXL, serves as a host factor for CHIKV. Furthermore, the phosphatidylserine binding domain of TIM-1, the so-called metal ion ligand binding site (MILIBS), is crucial for the host factor function of TIM-1 in the context of CHIKV infection. TIM-1 influences both the binding and internalisation of CHIKV particles to human cells. Finally, TIM-1 also enhances CHIKV infection in keratinocytes, which are among the first target cells of the virus after a mosquito bite. We therefore hypothesise that alphaviruses, including CHIKV, use phosphatidylserine receptors for cell attachment and entry into reservoir species such as non-human primates and into transmitting mosquito vectors. In addition, we want to clarify whether the virus produced in insect cells also releases phosphatidylserine and whether this is helpful in the infection of human cells, i.e. in cross-species transmission. And finally, we want to clarify how phosphatidylserine is exposed on the viral envelope.
This work will contribute to the understanding of the molecular composition of alphavirus particles, the function of alphavirus attachment factors and the role of apoptotic mimicry in cross-species transmission and consequently the emergence of alphaviruses.
Funding:
German Academic Exchange Service (DAAD), 48.000 EUR




